Architecture
This document is part of a series of documents describing the representation of metadata in BioPortal
The General Architecture For Representing Metadata in BioPortal
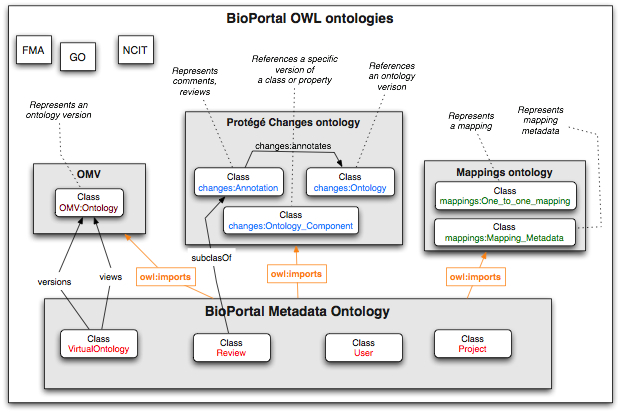
We describe the types of metadata that BioPortal has and the representation of this metadata in the BioPortal Metadata ontology. This ontology contains classes corresponding to the description of ontology metadata, mappings, reviews, marginal notes, projects, and users. The classes define specific properties for each type of metadata and the links between them.
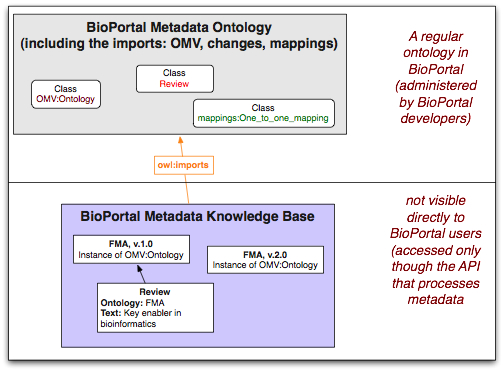
The concrete metadata that users contribute to BioPortal is represented in the BioPortal Metadata Knowledge Base (see the example in the figure). This knowledge base does not define any classes, it simply imports the BioPortal Metadata Ontology (the Architecture figure shows the overall architecture). The knowledge base contains instances of the classes defined in that ontology. When a service or another API call requests metadata information, the implementation queries the BioPortal Metadata Knowledge Base for the corresponding instance information.
We process the BioPortal Metadata ontology in exactly the same way as we process all other ontologies in BioPortal: it is represented in the BioPortal index, users can comment on it and reference it. The BioPortal Metadata Knowledge Base is also an OWL ontology, but it has a special status and BioPortal user interface does not directly display it in Bioportal. Instead, it queries it whenever it needs to access any metadata
The BioPortal Metadata Ontology ontology imports three other OWL ontologies:
- The Ontology Metadata Vocabulary (OMV) describes most of the metadata for ontologies themselves (e.g., domain, author, version number, ontology language, etc.)
- The Protégé Changes ontology provides the definitions for generic annotations (the Annotation class) and ontology components that they annotate.
- The Mappings ontology provides vocabulary for describing one-to-one mappings between concepts and corresponding metadata
The BioPortal Metadata ontology, by importing these three ontologies, can use all the classes and definitions from there and extend these definitions. The specific metadata is represented as instances of corresponding classes.
The BioPortal Metadata Ontology
The following ontologies are available for download and direct access at the Protege web site:
Classes and properties in the BioPortal Metadata ontology
We will now describe some key classes used to represent metadata.
Representing ontology metadata and provenance information
The metadata about ontologies themselves is represented mostly using the OMV vocabulary (namespace prefix “OMV”). OMV is the ontology developed jointly with the NeON consortium and accounts for much of the ontology metadata, including provenance information, static values (number of classes, etc.), licensing information, and so on.
The class OMV:Ontology
The main class in OMV is OMV:Ontology. It contains a set of metadata for a particular version of the ontology. Some properties of interest, among others, are:
- name
- acronym
- creationDate
- description (range: String)
- documentation (range: URL)
- endorsedBy (range: OMV:Party)
- one of the instances to fill in this value is OBOFoundry, an instance of OMV:Organisation
- domain
- ontologyLanguage
- keyClasses
- keywords
- ...
We also add some instances for ontology engineering tools and methodologies to account for OBO ontologies (OBO-Edit tool and the DAG ontology structure) and to include the OBO Foundry, caBIG, and others as endorsing organizations.
Full list of properties for instances of OMV:Ontology
Here is the full list of properties for instances of the class OMV:Ontology. It includes the properties that are added in the BioPortal metadata ontology, in addition to the ones that were inherited from OMV.
- designedForOntologyTask: a textual description of the task for which the ontology was designed
- endorsedBy: organizations that endorsed the ontology (e.g., caBIG, OBO Foundry, WHO, etc.)
- hasContributor: names of contributors to the ontology
- hasCreator: the creator of the ontology
- hasDomain: a category that describes the ontology, from a pre-defined list of categories (e.g., Anatomy, names of specific organisms, Diseases, etc.)
- hasFormalityLevel: is it a taxonomy, a formal ontology, etc.
- hasLicense: license under which the ontology is distributed
- hasOntologyLanguage: the language in which the ontology was developed
- usedOntologyEngineeringTool: the tool that was used to develop the ontology
- currentVersion: version number
- URI: the URI of the ontology
- acronym: the ontology acronym
- creationDate, modificationDate
- description: description of the ontology
- keyClasses: the list of key classes in the ontology
- keywords: the list of keywords describing the ontology
- name: name of the ontology
- status: alpha, pre-production. production, etc.
- hasContactEmail, hasContactEmail: contact information for the ontology authors (can be a mailing list)
- urlPublications: a link to the list of publications about the ontology
- urlHomepage: the homepage for the ontology (different from URI; this is a page intended for humans_
- synonymProperty: which property holds the synonyms for class names (default: skos:altLabel
- preferredNameProperty: which property holds the preferred names for classes (default: rdfs:label
- documentationProperty: which property holds the class definitions (default: skos:definition
- authorProperty: which property holds the author of a class (default: dc:creator
Administration and maintenance in BioPortal; database information for finding the ontology in BioPortal. These properties are specific to the BioPortal implementation
- administeredBy: the BioPortal user who administers the ontology
- codingScheme: used for UMLS terminologies
- fileNames: the name of the file for downloading the ontology
- filePath: the location of the file for downloading the ontology
- isFoundry: is the ontology pulled from the OBO Foundry site?
- oboFoundryID: the id of the ontology on the OBO Foundry site (used for OBO Pull)
- isManual: is ontology uploaded directly to BioPortal or pulled from a different repository (such as OBO Foundry)
- isRemote/tt>: is the ontology uploaded to BioPortal or did the authors provide only metadata, but not the ontology itself
- uploadDate: the date when the ontology was uploaded to BioPortal
Properties for handling ontology versioning:
- hasPriorVersion: a pointer to the earlier version of the ontology
- isVersionOfVirtualOntology: a pointer to the "virtual ontology" instance -- the container for all versions of the same ontology
- id: BioPortal internal version number
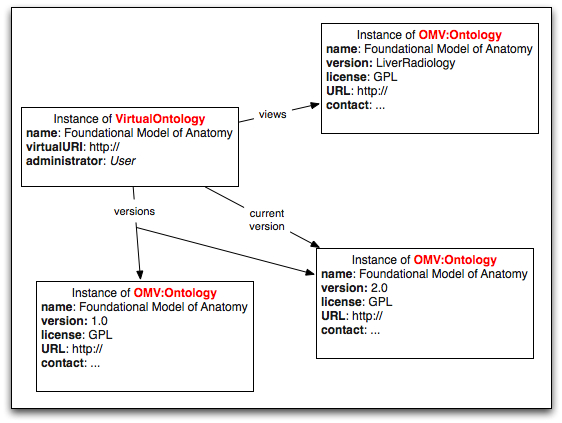
The class VirtualOntology
A class VirtualOntology collects the information about each virtual ontology in the repository, all its versions and views. There is one instance of the VirtualOntology class for each virtual ontology in BioPortal. Each such instance has the following properties:
- currentVersion (range: OMV:Ontology) is a pointer to the metadata describing the current version of the ontology;
- ontologyName (range: String) provides a name for the ontology (e.g., “NCI Thesaurus"); we don’t technically have to have it as we can get it from the current version, but it may be useful to have it here for convenience (but need to remember to update it)
- virtualURI (range: URI) provide a virtual URI for this ontology; it could also be an ontology ID, as used in BioPortal now
- hasView (range OMV:Ontology) provides a collection of instances of the OMV:Ontology class corresponding to the views of this virtual ontology
- hasVersion (range OMV:Ontology) provides a collection of instances of the OMV:Ontology class corresponding to the versions of this virtual ontology
The class Project
A class Project contains the information about each project. This information is represented as instances of the Project class. Each project has the following properties:
- projectName (range: String)
- description (range: String)
- institution (range: String)
- homepage (range: URL)
- administrator (range: User)
- hasContactInformation (range: String)
- usesOntologies (range: VirtualOntology)
The class BioPortalUser
A class BioPortalUser extends the class OMV:Person and foaf:Person. In addition, each user has the following properties:
- userName (range: String)
- createdReviews (range: Review)
- createdMarginalNotes (range: changes:Annotation)
- createdMappings (range: mappings:Mapping)
Representing marginal notes
Marginal notes and ontology reviews instances contain the content of the notes and reviews, their types, and the provenance information for them. The Protégé Changes ontology provides several key classes for representing ontology components and annotations and we extend these classes with several classes and properties relevant to BioPortal:
The classes and properties below describe annotations
The class changes:AnnotatableThing
The class changes:AnnotatableThing represents anything that can be annotated, including ontology components, ontologies themselves, mappings, other annotations, etc. This class has the following key property:
- changes:associatedAnnotations (range: changes:Annotation) links to all annotations that are associates with this object
The subclasses of changes:AnnotatableThing:
- changes:OntologyComponent (represents classes and properties; we will use this class to encapsulate how we address individual classes and properties)
- the class Ontology added in the BioPortal Metadata ontology to represent ontology as a whole as a target of a review
- changes:Annotation, includes marginal notes and reviews.
The class changes:Annotation
Instances of the class changes:Annotation represent reviews and marginal notes, as well as notes on mappings and other reviews. For now, marginal notes will be direct instances of Annotation because we do not distinguish between different types of marginal notes, such as questions or proposals. This class has the following key properties:
- changes:annotates, which is the inverse of the property changes:associatedAnnotations. This property has instances of changes:AnnotatableThing as its value. Thus, it refers to the exact version of a concept or an ontology for which annotation has been created.
- annotatesByVirtualID contains the virtual id for the ontology or the concept that this annotation annotates. We need this property for efficient retrieval so that we can query by the id.
We will use two subclasses of the class Annotation:
- the class Review that represents the reviews of the ontologies (see below).
- the class Proposal (and its subclasses) to represent structured proposals
The class changes:Proposal
Many of the structured notes in BioPortal are proposals for ontology changes. We represent several types of change proposals:
- New term proposal (class changes:NewEntityProposal): a structured proposal to create a new term in an ontology. It contains the following fields:
- Generated ID (property changes:entityID): an automatically generated id for the new term that the proposer can use while the ontology curators are adding the term to the ontology
- Preferred name (property changes:preferredName): preferred name (label) for the new term
- Synonyms (property changes:synonyms): synonyms for the new term
- Definition (property changes:definition): proposed definition for the new term
- Superclass (property changes:parent): proposed parents of the new term (the note is usually attached to the proposed parent; so, by default the value for changes:parent is the same as the value for changes:annotates)
- Comment (property changes:reasonForChange): a free-text comment explaining the reason for the proposed change
- New hierarchical relationship proposal (class changes:ChangeHierarchyProposal): a proposal to change the location of the class in the hierarchy. It contains the following fields:
- Hierarchical relationship (property changes:relationType): The hierarchical relationship that needs to be changed (e.g., part-of or subclass-of)
- Old parent (property changes:oldParents): the old parent in the hierarchy (the proposal is usually attached to the old parent; so, by default the value for changes:oldParents is the same as the value for changes:annotates)
- New parent (property changes:newParent): where the class should be moved
- Keep the old parent? (property changes:moveOrCopy): should the proposed new parent be added to the list of parents (copy) or should the old parent be removed and the class should be moved to the new parent (move)?
- Change property value proposal (class changes:PropertyValueChangeProposal): change a value for a property or create a new value. Contains the following fields:
- New value (property changes:newValue): the new/added value for the property
- Old value (property changes:oldValue): the property value the new value will be replacing (if any; if this property is false, then it is a proposal to add a new value, rather than replace an old one).
- Retire a concept (class changes:RetireProposal): proposal to retire the concept to which it is attached
- Split a concept into two concepts (class changes:SplitProposal): a proposal to split the concept to which this note is attached into two concepts. The fields are:
- Generated ID (property changes:entityID): an automatically generated id for the new term that the proposer can use while the ontology curators are adding the term to the ontology
- Preferred name (property changes:preferredName): preferred name (label) for the new term
Note: changes:SplitProposal will probably be a subclass of changes:NewEntityProposal.
- Merge this concept with another concept (class changes:MergeProposal): Merge the selected concept into another concept. The expected side-effect is that the current concept will be retired (Thus, changes:MergeProposal may need to be a subclass of changes:RetireProposal). The fields are:
- Merge with (property changes:targetEntity): what to merge the current entity with.
Representing reviews
The class bpMetadata:Review
A class bpMetadata:Review is a subclass of changes:Annotation. This class represents reviews of an ontology as a whole. Each review contains ratings and comments along each of the dimensions specified as the evaluation dimensions (class EvaluationDimension, with two properties: numericRating, textualReview). It has the following properties, in addition to the provenance ones inherited from the Annotation class:
- inTheContextOfProject (range: Project; inverse property: ontologyReviews) indicates that a review was created in the context of a particular project
- the range for the property annotates is restricted to OMV:Ontology
- A property for each of the review dimensions. These properties are all subproperties of the property reviewOnDimension (range: EvaluationDimension (see figure)):
- digreeOfFormalityReview
- documentationAndSupportReview
- usabilityReview
- domainCoverageReview
- correctnessReview
- qualityOfContentReview
This list of properties can be changed or extended, and, thus, other implementations can custom-tailor the dimensions. Each property has an annotation property rdfs:label that contains a string that will be displayed for this dimension (e.g., "Domain coverage"). If the order in which these dimensions appear is important, the developers will need to update the Constant Names.DIMENSIONS in the code.
Representing Mappings
The Mappings ontology represents different types of mappings and the corresponding metadata. More details on the mapping metadata in a separate document. We define the mapping ontology for BioPortal to represent one-to-one mappings. This ontology has the key classes described below.
The class mappings:One_to_one_mapping
A class mappings:One_to_one_mapping represents a mapping between any two concepts (usually, from different ontologies) It has the following properties (all in the mappings namespace:
- source (range: URL)
- target (range: URL)
- relation (range: URL)
- mapping_metadata (range: Mapping_Metadata)
In practice, for each source and target, we will have a property for:
- virtual ontology id
- ontology version id
- full concept id
Note: Currently mappings are stored using the short id for the concept. We will need to replace the short ids with full ids when we store the mappings in the back end.
The class mappings:Mapping_Metadata
A class mappings:Mapping_Metadata represents the metadata about each mapping:
- applicationContext (range: String)
- authority (range: Authority, a class with two subclasses Authority_Algorithm and Authority_Manual; the former contains the details of the algorithm)
- comment (range: String)
- date (range: date)
- dependency (range: One_to_one_mapping)
- externalReferences (range: String)
- submittedBy (range: undefined)
- verified (range: undefined)
Note that we change the range for mappings:source and mappings:target to be changes:Ontology_Component. Thus, we will re-use whatever solution we choose to represent targets of annotations. Because one can also add comments to mappings, we add changes:AnnotatableThing as another superclass of the class One_To_One_Mapping. Thus, One_To_One_Mapping now has changes:associatedAnnotations as one of its properties.
Representing Metrics
Please refer to the Ontology Metrics page for the details of metrics representation in BioPortal.
Representing Views
We treat a view as an ontology and therefore the representation of views parallels the representation of ontologies themselves.
The representation described here is flexible enough to address all the requirements that we identified. While the representation is somewhat complex to allow the full flexibility (views on views, versions of views, etc.), the user interface will hide most of the complexity.
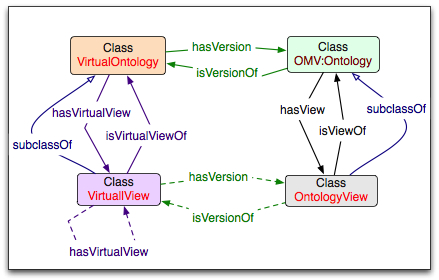
Just as we have a VirtualOntology class as a container for all versions of a particular ontology, we have a class VirtualView to represent a container of all versions of a particular view. For example, we can have a "Liver" view of the FMA. The instance of the VirtualView class will point to all specific versions of the view. These specific versions are instances of the class OntologyView, which is a subclass of the OMV:Ontology class. Each instance of OntologyView (i.e., a specific version of the view), points to the specific version of the "base" ontology that was used to create this view.
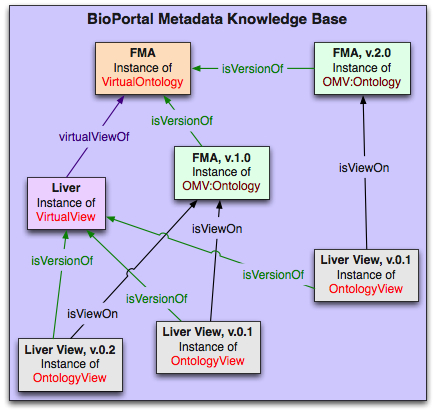
The two figures to the right show [#viewClasses|the key classes] in representing the views, and an example of [#viewInstances|a set of instances] that instantiate these classes. The color of the box for the instance in the second figure is the same as the color of its class in the first figure.
There is a VirtualView instance that contains the collections of all the versions of this view. Some of the versions of the view were created using v.1.0 of the FMA. One version of the Liver view was created using v2.0 of the FMA.
The class OntologyView
The class OntologyView is the main class that represents the metadata about a specific version of the view. This class is a subclass of the class OMV:Ontology, which represents the metadata about a specific version of an ontology. Any specific version of a view is fundamentally just an ontology; thus it inherits all the metadata properties of the OMV:Ontology class (copied below for convenience):
- name
- acronym
- creationDate
- description (range: String)
- documentation (range: URL)
- endorsedBy (range: OMV:Party)
- one of the instances to fill in this value is OBOFoundry, an instance of OMV:Organisation
- domain
- ontologyLanguage
- keyClasses
- keywords
In addition to inheriting all the metadata properties from that describe all BioPortal ontologies, we need to provide additional metadata for each version of a view:
- viewDefinition (range String) has the textual representation of the view definition. This definition can be a query that was used to create the view, a set of traversal directive (as in Prompt), or any other way that specifies how the view was extracted. (Question: is the type String sufficient here or will we need something more structured?)
- viewDefinitionLanguage (range: ViewDefinitionLanguage) defines the language that was used to define the view (e.g., the query language, such as SPARQL, for a view that was generated by a query). The instance of the class ViewDefinitionLanguage will have all the pertinent attribute of the language (name, creator, url) as well as the specific version that was used for the view.
- viewGenerationEngine (range: ViewGenerationEngine) defines the engine that was used to compute the view. The instance of the class ViewGenerationEngine will have all the pertinent attribute of the engine (name, creator, url) as well as the specific version that was used to generate the view.
The class VirtualView
The class VirtualView collects the information about all the versions of a particular view. This class is a subclass of the VirtualOntology class. Thus, it inherits all of its properties. However, we restrict the range of values for the properties currentVersion, hasView and hasVersion to instances of the OntologyView class. Here is the summary of the inherited properties for the class VirtualView, with the corresponding restrictions:
- currentVersion (range: OntologyView) is a pointer to the metadata describing the current version of the view;
- ontologyName (range: String) provides a name for the view (e.g., “Liver view for radiology")
- virtualURI (range: URI) provide a virtual URI for this view; it could also be an ontology ID, as used in BioPortal now
- hasView (range OntologyView) provides a collection of instances of the OntologyView class corresponding to the views of this view (i.e., views on views)
- hasVersion (range OntologyView) provides a collection of instances of the OntologyView class corresponding to the versions of this virtual view
There is also one additional property defined for the class VirtualView:
- isVirtualViewOf (range: VirtualOntology) is a pointer to the "base" ontology or ontologies from which the view is created (e.g., FMA). Note that we may later decide not to use this property as there may be a situation as the set of "base" ontologies is different for different versions of the view. For instance, one version is a view on FMA; but then we extend this version and join it with some classes from NCI Thesaurus. If we want to allow for such evolution, then we don't want to store the "base" ontology (or ontologies) at the level of the VirtualView. We can always infer this information from the specific versions that we access through the property hasVersion.