OboInOwl:Main Page
OboInOwl
This wiki is for discussing the mapping between Obo1.2 format and OWL
And you can find the first version of this mapping here:
http://www.godatabase.org/dev/mapping-obo-to-owl.html
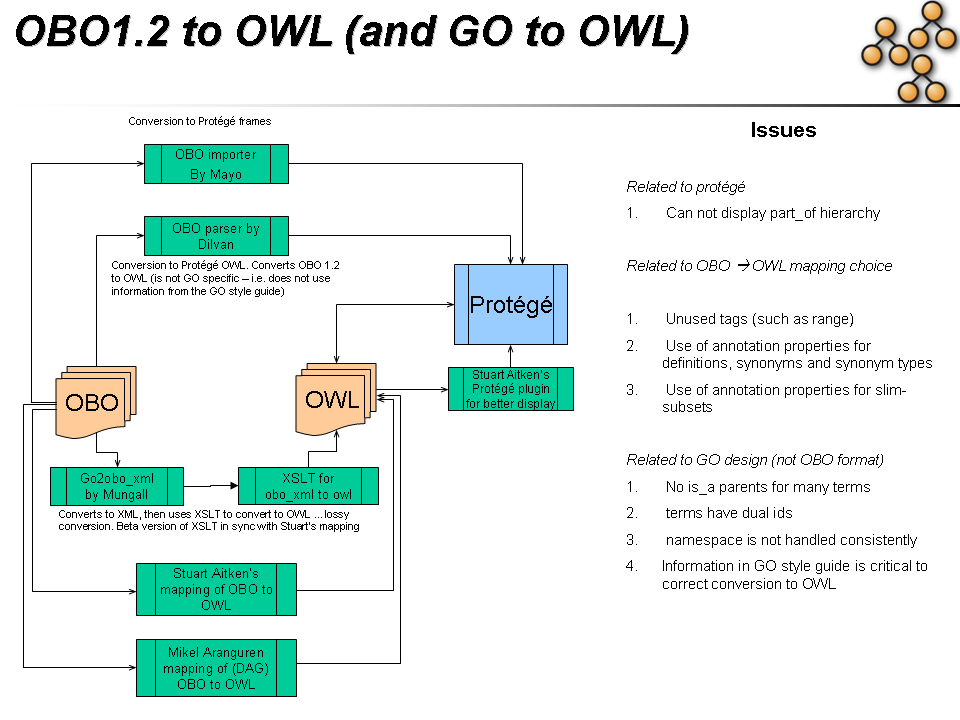
We are working on a newer version of this mapping
Mail Lists
Also of interest:
Progress
I've overhauled the obo2owl mapping. I've pretty much followed Alan's recommendations (I made a lot of purely internal changes to the xslt too though which should make it much clearer). Hope these work for you Stuart. Sorry about the churn - but this will definitely be worth it in the end.
Example OWL file can be found here (also attached):
http://geneontology.cvs.sourceforge.net/*checkout*/geneontology/go-dev/xml/examples/gotest.owl
(note that this example includes a cross-product example)
The OWL is generated from either of the following:
http://geneontology.cvs.sourceforge.net/*checkout*/geneontology/go-dev/xml/examples/gotest.obo http://geneontology.cvs.sourceforge.net/*checkout*/geneontology/go-dev/xml/examples/gotest.obo-xml
The XSL can be found here:
http://geneontology.cvs.sourceforge.net/*checkout*/geneontology/go-dev/xml/xsl/obo2owl.xsl http://geneontology.cvs.sourceforge.net/*checkout*/geneontology/go-dev/xml/xsl/obo2owl_obo_in_owl_metamodel.xsl
The XSL actually serves as fairly reasonable documentation about what's going on - but we'll also come up with a friendlier description once it's finalised
You can convert the obo-xml directly with the xslt. If you want to convert from obo you'll need the latest version of go-perl (from cvs)
Here are the changes and things still pending:
Adopted Alan Ruttenberg's metamodel changes (see obo-format list)
split into 2 separate xsl files
subset (ontology views) now more consistent with obo
* the oboInOwl class is SubsetDef
* this does not appear in the owl:Ontology section, it stands alone
(subsets can be used across ontologies)
namespace changes -
* the metamodel is now called oboInOwl
(the format is owned by GO, so this maps to a GO URI)
* the default ontology content namespace is now bioont
(the URI for this will be some bioontologies.org URI)
* slashes not hashes or underscores
- example: rdf:about="oboContent/GO/0000001"
fixed rdf:about/resource/ID issues
- ID is never used
- about and resource now used in correct places
CHECKED
- validates as DL in http://phoebus.cs.man.ac.uk:9999/OWL/Validator - works in SWOOP - works in Protege-OWL (but looks odd)
TODO
do we need an equivalentClass for intersectionOf?
SWOOP saves this without
decide on final URI scheme
- Can we make the URIs less verbose? Use entities - or is this frowned on?
new obo tags for obsoletion
handling obsoletes
decide on whether the oboInOwl metamodel should be exported as
part of the content export, or linked to separately;
and if linked to separately, do we need an owl:imports?
Overview of [Other] Mapping efforts
Below is a quick summary of the various OBO to OWL conversion efforts that I am aware of. Email me (nigam .AT. stanford.edu) with additions/deletions as you come across them.