Difference between revisions of "SKOSSupport"
(added concrete example) |
|||
| (14 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[File:Plos_thesaurus_class_tree.png|thumb|400px|PLOS Thesaurus concept tree in BioPortal]] | [[File:Plos_thesaurus_class_tree.png|thumb|400px|PLOS Thesaurus concept tree in BioPortal]] | ||
| − | |||
| − | |||
__TOC__ | __TOC__ | ||
| − | |||
=== Support for SKOS vocabularies in BioPortal === | === Support for SKOS vocabularies in BioPortal === | ||
| Line 23: | Line 20: | ||
<nowiki><http://www.example.com/animals></nowiki> rdf:type skos:Concept | <nowiki><http://www.example.com/animals></nowiki> rdf:type skos:Concept | ||
| − | In SKOS vocabularies, BioPortal only treats the SKOS concept assertions as concepts to be displayed. If the vocabulary contains other assertions about other types of concepts, BioPortal will not treat these as concepts in any of its displays or features. See the W3C's SKOS System Primer and SKOS Reference for concept documentation and examples: | + | In SKOS vocabularies, BioPortal only treats the SKOS concept assertions as concepts to be displayed. If the vocabulary contains other assertions about other types of concepts, BioPortal will not treat these as concepts in any of its displays or features. |
| + | |||
| + | See the W3C's SKOS System Primer and SKOS Reference for concept documentation and examples: | ||
https://www.w3.org/TR/2009/NOTE-skos-primer-20090818/#secconcept | https://www.w3.org/TR/2009/NOTE-skos-primer-20090818/#secconcept | ||
https://www.w3.org/TR/skos-reference/#concepts | https://www.w3.org/TR/skos-reference/#concepts | ||
| + | |||
| + | '''Note:''' Some OWL ontologies declare the SKOS namespace to facilitate minimal use of SKOS constructs for things like labels (e.g., skos:prefLabel, skos:altLabel) or mappings (e.g., skos:exactMatch, skos:broaderMatch). In these cases, the proper format for new ontology submissions is OWL, not SKOS. | ||
| + | |||
===== skos:ConceptScheme & skos:hasTopConcept ===== | ===== skos:ConceptScheme & skos:hasTopConcept ===== | ||
| Line 51: | Line 53: | ||
https://www.w3.org/TR/skos-reference/#schemes | https://www.w3.org/TR/skos-reference/#schemes | ||
| − | ===== skos:broader | + | If your vocabulary declares more than one concept scheme, all of the top concepts will be aggregated and displayed as root level concepts. BioPortal's user interface doesn't provide support for grouping top level concepts by concept scheme. It is recommended to declare a owl:Ontology, especially for metadata annotations. |
| + | |||
| + | |||
| + | ==== Hierarchy in SKOS vocabularies ==== | ||
| + | |||
| + | The only semantic relationship in SKOS vocabularies that BioPortal uses to construct and display concept hierarchies is the skos:broader property. | ||
| + | |||
| + | ex:mammals rdf:type skos:Concept; | ||
| + | skos:prefLabel "mammals"@en; | ||
| + | skos:broader ex:animals. | ||
| + | |||
| + | Other properties used to denote hierarchical relationships like skos:narrower, skos:broaderTransitive, and skos:narrowerTranstive, are ignored. | ||
| + | |||
| + | |||
| + | ==== Metrics data for SKOS vocabularies ==== | ||
| + | |||
| + | BioPortal uses the [https://owlcs.github.io/owlapi/ OWL API] for parsing all ontology and vocabulary submissions, as well as for the calculation of metrics data. The OWL API treats SKOS vocabularies as RDF files containing classes and instances. According to the SKOS Reference, concepts are [https://www.w3.org/TR/skos-reference/#L842 instances of owl:Class], and thus are counted as instances (a.k.a. "individuals"). | ||
| + | |||
| + | When viewing metrics tables in the BioPortal user interface, the value for the "NUMBER OF INDIVIDUALS" corresponds to the number of concepts in any given SKOS vocabulary. | ||
| + | |||
| + | |||
| + | [[File:Metrics.png|none|frame|PLOS Thesaurus metrics in BioPortal]] | ||
| + | |||
| + | |||
| + | ==== SKOS-XL ==== | ||
| + | |||
| + | Currently BioPortal offers no support for the [https://www.w3.org/TR/skos-reference/#xl SKOS eXtension for Labels (SKOS-XL)]. A suggested workaround for SKOS vocabularies that make use of SKOS-XL, is to dump the value of labels (i.e., skosxl:literalForm of skosxl:*Label instances) into the corresponding skos:*Label property. | ||
| + | |||
| + | |||
| + | ==== SKOS mapping properties ==== | ||
| + | |||
| + | At this time, BioPortal doesn't use SKOS mapping properties, i.e., skos:*Match, to populate the mapping repository. One-to-one mappings between SKOS concepts need to be uploaded separately via the [http://data.bioontology.org/documentation BioPortal REST API]. | ||
| + | |||
| + | === Example of valid SKOS === | ||
| + | |||
| + | This example provides a simple illustration of the composition of a SKOS file that complies with the above constraints. | ||
| + | |||
| + | ==== Example header ==== | ||
| + | |||
| + | The header shown here defines a few typical namespaces that may be useful. | ||
| + | |||
| + | The last namespace is the one that defines this SKOS vocabulary. Ideally, the IRI defining the myskosid namespace is the resolvable location of the SKOS ontology. | ||
| + | |||
| + | <?xml version="1.0" encoding="UTF-8"?> | ||
| + | <rdf:RDF | ||
| + | xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#" | ||
| + | xmlns:rdfs="http://www.w3.org/2000/01/rdf-schema#" | ||
| + | xmlns:skos="http://www.w3.org/2004/02/skos/core#" | ||
| + | xmlns:dct="http://purl.org/dc/terms/" | ||
| + | xmlns:myskosid="https://example.org/ontologies/myskosontology/" | ||
| + | > | ||
| + | |||
| + | ==== Example ontology description ==== | ||
| + | |||
| + | In the rdf:type item, this namespace is declared as the ConceptScheme. The ConceptScheme does not have to be the same as the namespace of the ontology. | ||
| + | |||
| + | Other metadata is provided as an example of good practices in ontology metadata. The dct:creator does not have to be an ORCID ID, but a unique identifier is an ideal way of naming a creator (whether individual or organization). | ||
| + | |||
| + | This ontology has only 2 concepts (to be defined below), hence only 2 skos:hasTopConcept declarations. | ||
| + | |||
| + | <rdf:Description rdf:about="https://example.org/ontologies/myskosontology/"> | ||
| + | <rdfs:label xml:lang="en">Example SKOS ontology for BioPortal</rdfs:label> | ||
| + | <rdf:type rdf:resource="http://www.w3.org/2004/02/skos/core#ConceptScheme"/> | ||
| + | <rdfs:comment xml:lang="en">Example created to simplify understanding and creation of a SKOS vocab for BioPortal</rdfs:comment> | ||
| + | <dct:created rdf:datatype="http://www.w3.org/2001/XMLSchema#date">2020-09-16</dct:created> | ||
| + | <dct:modified rdf:datatype="http://www.w3.org/2001/XMLSchema#date">2020-09-16</dct:modified> | ||
| + | <dct:license rdf:resource="https://creativecommons.org/licenses/by/4.0/"/> | ||
| + | <dct:creator rdf:resource="https://orcid.org/0000-0001-6875-5360"/> | ||
| + | <skos:hasTopConcept rdf:resource="https://example.org/ontologies/myskosontology/fragmentid001"/> | ||
| + | <skos:hasTopConcept rdf:resource="https://example.org/ontologies/myskosontology/fragmentid002"/> | ||
| + | </rdf:Description> | ||
| + | |||
| + | ==== Example term definitions ==== | ||
| + | |||
| + | This section shows the two concepts and a few typical annotations about those concepts. The first rdf:Description line of each group names the concept that is being defined in the indented lines following. | ||
| + | |||
| + | The rdf:Type and skos:prefLabel are required annotation content for BioPortal to work effectively. Other items are optional. | ||
| + | |||
| + | The skos:topConceptOf is not strictly required for BioPortal SKOS ontologies, but provides useful contextualization if there is more than one topConcept. | ||
| + | |||
| + | <rdf:Description rdf:about="https://example.org/ontologies/myskosontology/fragmentid001"> | ||
| + | <rdf:type rdf:resource="http://www.w3.org/2004/02/skos/core#Concept"/> | ||
| + | <skos:prefLabel xml:lang="en">First concept</skos:prefLabel> | ||
| + | <skos:definition xml:lang="en">The very first example provided as part of this ontology.</skos:definition> | ||
| + | <skos:topConceptOf rdf:resource="https://example.org/ontologies/myskosontology/"/> | ||
| + | </rdf:Description> | ||
| + | <rdf:Description rdf:about="https://example.org/ontologies/myskosontology/fragmentid002"> | ||
| + | <rdf:type rdf:resource="http://www.w3.org/2004/02/skos/core#Concept"/> | ||
| + | <skos:prefLabel xml:lang="en">Second Concept</skos:prefLabel> | ||
| + | <skos:definition xml:lang="en">The second example provided as part of this ontology.</skos:definition> | ||
| + | <skos:topConceptOf rdf:resource="https://example.org/ontologies/myskosontology/"/> | ||
| + | </rdf:Description> | ||
| + | |||
| + | ==== Closing XML ==== | ||
| − | + | Needed for a complete, parseable RDF file! | |
| − | |||
| − | + | </rdf:RDF> | |
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 03:17, 18 September 2020
Support for SKOS vocabularies in BioPortal
BioPortal is a web-based portal for accessing and sharing ontologies. The application accepts ontology submissions in OWL and OBO format, and SKOS vocabularies that contain particular constructs.
This wiki page documents the minimum set of SKOS constructs that must be present in a SKOS vocabulary for BioPortal to accept and handle the submission properly.
Please note that the SKOS constructs described here are handled only for vocabularies that are identified as SKOS when they are submitted to BioPortal. Vocabularies submitted as OWL or OBO formats are not examined for SKOS constructs.
Required SKOS constructs
skos:Concept
Concepts are the fundamental elements of SKOS vocabularies and are asserted using the skos:Concept class, e.g.:
<http://www.example.com/animals> rdf:type skos:Concept
In SKOS vocabularies, BioPortal only treats the SKOS concept assertions as concepts to be displayed. If the vocabulary contains other assertions about other types of concepts, BioPortal will not treat these as concepts in any of its displays or features.
See the W3C's SKOS System Primer and SKOS Reference for concept documentation and examples:
https://www.w3.org/TR/2009/NOTE-skos-primer-20090818/#secconcept
https://www.w3.org/TR/skos-reference/#concepts
Note: Some OWL ontologies declare the SKOS namespace to facilitate minimal use of SKOS constructs for things like labels (e.g., skos:prefLabel, skos:altLabel) or mappings (e.g., skos:exactMatch, skos:broaderMatch). In these cases, the proper format for new ontology submissions is OWL, not SKOS.
skos:ConceptScheme & skos:hasTopConcept
For every ontology entry in BioPortal, the application provides a tabbed interface with various views of the ontology data, e.g., a "Classes" tab with a tree structure to graphically depict the hierarchical collection of ontology classes.
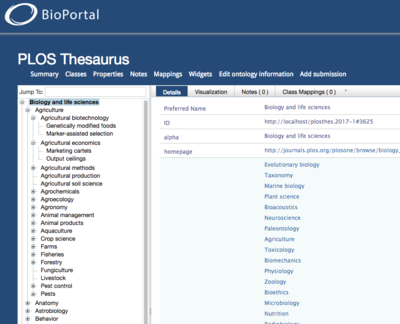
In the case of SKOS vocabularies, BioPortal determines which concepts to display as roots in the concept tree by querying vocabulary content for occurrences of skos:hasTopConcept property assertions. Top concepts are the most general concepts contained in SKOS concept schemes (an aggregation of one or more SKOS concepts).
The following example, taken from the SKOS System Primer, shows how to define a concept scheme and link it to the most general concepts it contains:
@prefix skos: <http://www.w3.org/2004/02/skos/core#> . @prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> . @prefix ex: <http://www.example.com/> . ex:animalThesaurus rdf:type skos:ConceptScheme; skos:hasTopConcept ex:mammals; skos:hasTopConcept ex:fish.
SKOS vocabularies submitted to BioPortal must contain a minimum of one concept scheme and top concept assertion. See the the SKOS System Primer and SKOS Reference for more documentation of concept schemes and top concepts:
https://www.w3.org/TR/2009/NOTE-skos-primer-20090818/#secscheme
https://www.w3.org/TR/skos-reference/#schemes
If your vocabulary declares more than one concept scheme, all of the top concepts will be aggregated and displayed as root level concepts. BioPortal's user interface doesn't provide support for grouping top level concepts by concept scheme. It is recommended to declare a owl:Ontology, especially for metadata annotations.
Hierarchy in SKOS vocabularies
The only semantic relationship in SKOS vocabularies that BioPortal uses to construct and display concept hierarchies is the skos:broader property.
ex:mammals rdf:type skos:Concept; skos:prefLabel "mammals"@en; skos:broader ex:animals.
Other properties used to denote hierarchical relationships like skos:narrower, skos:broaderTransitive, and skos:narrowerTranstive, are ignored.
Metrics data for SKOS vocabularies
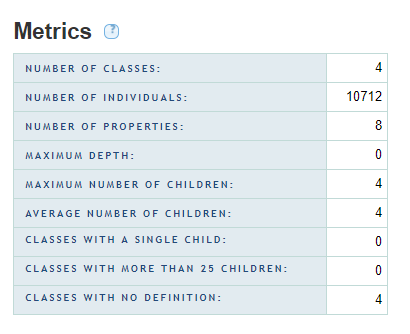
BioPortal uses the OWL API for parsing all ontology and vocabulary submissions, as well as for the calculation of metrics data. The OWL API treats SKOS vocabularies as RDF files containing classes and instances. According to the SKOS Reference, concepts are instances of owl:Class, and thus are counted as instances (a.k.a. "individuals").
When viewing metrics tables in the BioPortal user interface, the value for the "NUMBER OF INDIVIDUALS" corresponds to the number of concepts in any given SKOS vocabulary.
SKOS-XL
Currently BioPortal offers no support for the SKOS eXtension for Labels (SKOS-XL). A suggested workaround for SKOS vocabularies that make use of SKOS-XL, is to dump the value of labels (i.e., skosxl:literalForm of skosxl:*Label instances) into the corresponding skos:*Label property.
SKOS mapping properties
At this time, BioPortal doesn't use SKOS mapping properties, i.e., skos:*Match, to populate the mapping repository. One-to-one mappings between SKOS concepts need to be uploaded separately via the BioPortal REST API.
Example of valid SKOS
This example provides a simple illustration of the composition of a SKOS file that complies with the above constraints.
Example header
The header shown here defines a few typical namespaces that may be useful.
The last namespace is the one that defines this SKOS vocabulary. Ideally, the IRI defining the myskosid namespace is the resolvable location of the SKOS ontology.
<?xml version="1.0" encoding="UTF-8"?> <rdf:RDF xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#" xmlns:rdfs="http://www.w3.org/2000/01/rdf-schema#" xmlns:skos="http://www.w3.org/2004/02/skos/core#" xmlns:dct="http://purl.org/dc/terms/" xmlns:myskosid="https://example.org/ontologies/myskosontology/" >
Example ontology description
In the rdf:type item, this namespace is declared as the ConceptScheme. The ConceptScheme does not have to be the same as the namespace of the ontology.
Other metadata is provided as an example of good practices in ontology metadata. The dct:creator does not have to be an ORCID ID, but a unique identifier is an ideal way of naming a creator (whether individual or organization).
This ontology has only 2 concepts (to be defined below), hence only 2 skos:hasTopConcept declarations.
<rdf:Description rdf:about="https://example.org/ontologies/myskosontology/"> <rdfs:label xml:lang="en">Example SKOS ontology for BioPortal</rdfs:label> <rdf:type rdf:resource="http://www.w3.org/2004/02/skos/core#ConceptScheme"/> <rdfs:comment xml:lang="en">Example created to simplify understanding and creation of a SKOS vocab for BioPortal</rdfs:comment> <dct:created rdf:datatype="http://www.w3.org/2001/XMLSchema#date">2020-09-16</dct:created> <dct:modified rdf:datatype="http://www.w3.org/2001/XMLSchema#date">2020-09-16</dct:modified> <dct:license rdf:resource="https://creativecommons.org/licenses/by/4.0/"/> <dct:creator rdf:resource="https://orcid.org/0000-0001-6875-5360"/> <skos:hasTopConcept rdf:resource="https://example.org/ontologies/myskosontology/fragmentid001"/> <skos:hasTopConcept rdf:resource="https://example.org/ontologies/myskosontology/fragmentid002"/> </rdf:Description>
Example term definitions
This section shows the two concepts and a few typical annotations about those concepts. The first rdf:Description line of each group names the concept that is being defined in the indented lines following.
The rdf:Type and skos:prefLabel are required annotation content for BioPortal to work effectively. Other items are optional.
The skos:topConceptOf is not strictly required for BioPortal SKOS ontologies, but provides useful contextualization if there is more than one topConcept.
<rdf:Description rdf:about="https://example.org/ontologies/myskosontology/fragmentid001"> <rdf:type rdf:resource="http://www.w3.org/2004/02/skos/core#Concept"/> <skos:prefLabel xml:lang="en">First concept</skos:prefLabel> <skos:definition xml:lang="en">The very first example provided as part of this ontology.</skos:definition> <skos:topConceptOf rdf:resource="https://example.org/ontologies/myskosontology/"/> </rdf:Description> <rdf:Description rdf:about="https://example.org/ontologies/myskosontology/fragmentid002"> <rdf:type rdf:resource="http://www.w3.org/2004/02/skos/core#Concept"/> <skos:prefLabel xml:lang="en">Second Concept</skos:prefLabel> <skos:definition xml:lang="en">The second example provided as part of this ontology.</skos:definition> <skos:topConceptOf rdf:resource="https://example.org/ontologies/myskosontology/"/> </rdf:Description>
Closing XML
Needed for a complete, parseable RDF file!
</rdf:RDF>