Virtual Appliance FAQ
Ontology Management
How do I identify version numbers for ontologies stored in the system?
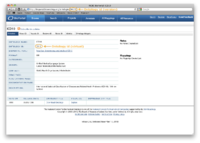
NCBO Ontology Web Services can be used with either the ontology identifier (sometimes referred to as the virtual identifier) or the ontology version identifier (sometimes referred to as the version id). The ontology identifier is used to identify and ontology and all of its versions. The ontology version identifier represents a single ontology version. The following screenshot shows how to identify these version numbers using the BioPortal Web UI.
How do I add or change categories?
- Download the Protégé client (Version Protégé 3.4.4)
- Download a copy of the BioPortal Metadata Protégé project file and modify as follows
- Line 639: make sure this matches your BioPortal Ontology Services database (ip address or domain of the virtual appliance)
- Line 649: this should be your MySQL username (bp_protege)
- Line 654: this should be your MySQL password (bioportalprotege)
- Note: These are the default username/password which should be changed for security purposes. If you have already changed them, please use the new username/password.
- Open the project file using Protégé
- Go to the "Individuals" tab
- Look for OMV:OntologyDomain and click it
- A list of "Asserted Instances" should show up, these are the defaults
- You can create or delete instances by clicking on the buttons
- Instances should have the following properties:
- id = unique integer value
- omv:name = display name for the category
- omv:isSubDomainOf = you can create hierarchies by making categories subDomains of one another
How can I migrate ontologies from BioPortal or previous NCBO Virtual Appliance versions into a new Appliance?
NCBO developers have created a script that can be used to automatically import ontologies from BioPortal or previous versions of the Virtual Appliance. The script was written in Ruby and is included on the Appliance. To use the script, do the following (requires knowledge of Linux command-line tools):
- Edit the script, located here:
/var/rails/BioPortal/current/util/import_ontologies/import_ontologies.rb- Make sure you have valid API keys for the FROM and TO systems.
- Assign the $FROM and $TO variables and ports as appropriate. The default configuration points to NCBO's BioPortal but can be changed to the location of your Virtual Appliance instance.
- The $ONTOLOGY_OWNER is the user id of the user who should own the ontologies on the TO system.
- $INCLUDE_ONTOLOGIES can be uncommented and used to import specific ontologies only. Otherwise all ontologies from the FROM system will be imported.
- Save your edits and continue below.
- Run the script using this command:
sh /var/rails/BioPortal/current/util/import_ontologies/import_ontologies.rb - The script will take some time to complete, updating the console as it runs. If automatic parsing is enabled, the ontologies should get parsed when the process next runs.
Please Note: The import script will attempt to use category and group IDs assigned to ontologies on the FROM system if they exist on the TO system as well. Ontology artifacts, such as notes, views, and mappings, are not imported. Only the most recent version of ontologies are imported; there is no method to import multiple versions of the same ontology using a common virtual id.
Ontology Parsing
When are new ontologies parsed?
The BioPortal Ontology Services application uses a scheduler to run a process that collects newly submitted ontologies and parses them, adds them to the search index, and calculates metrics. You can also parse ontologies manually, but you will need to index them for search and calculate metrics manually to have that information available.
The default schedule can be disabled by doing the following:
- Open /ncbo/sources/bioportal/tags/1030/build.properties
- Look for the section that starts with "# Ontology Parse Scheduler properties"
- Set
ontology.parse.scheduler.enabledto 'false' - From the /ncbo/sources/bioportal/tags/1030/ directory, run 'ant clean deploywar'
How do I manually parse an ontology?
To manually parse an ontology, visit the admin interface at http://example:8080/bioportal_admin and select "Parse Ontologies" from the list at the right. You must enter the version id, or a comma-separated list of ids, and then click "Run". The process will return with errors if it encounters any.
How do I know if an ontology has parsed?
Because the BioPortal Web UI uses aggressive caching, it may not immediately reflect the status of an ontology once it's been submitted. By default the ontology status on the Web UI is updated once every four or 12 hours (depending where you are viewing the information). You can change these default options by doing the following:
- Open /var/rails/BioPortal/current/app/models/data_access.rb
- Change the following in the getOntologyList method:
return self.cache_pull("ont_list", "getOntologyList", nil, MEDIUM_CACHE_EXPIRE_TIME)return self.cache_pull("ont_list", "getOntologyList", nil, 60*15)
- Change the following in the getOntology method:
return self.cache_pull("#{ontology_id}::_details", "getOntology", { :ontology_id => ontology_id })return self.cache_pull("#{ontology_id}::_details", "getOntology", { :ontology_id => ontology_id }, 60*15)
- This will change the Web UI so that it refreshed information about ontologies every 15 minutes
- You will need to run /sbin/service httpd restart to have the change take affect
- NOTE: You will need to make this change again if you update the Web UI code. In addition, if you have a large list of ontologies this could slow down the Web UI for users as it will be required to retrieve more information more often.
In addition, you can look at the REST service directly, which will always give you the most updated information. To do this, visit the following URL:
- http://example:8080/bioportal/ontologies/{ontology_version_id}
- You can look for the statusId element to get the status, the codes are as follows:
- 1 - Waiting
- 2 - Parsing
- 3 - Ready
- 4 - Error
- 5 - Not Applicable (for metadata-only ontologies)
Is there a log file for parsing?
Parsing progress is logged in the BioPortal Ontology Services log files.
There is a separate log file for OBO ontologies that can be monitored:
tail -f /var/log/tomcat6/lexgrid/LexBIG_load_log.txt
OWL and Protege-based ontologies are logged into the general log file:
tail -f /var/log/tomcat6/bioportal.log
How can I manually remove the hierarchy information from an ontology like NCI Metathesaurus?
- Find the database prefix of the ontology by looking up the ontology in the LexEVS registry.xml file.
- Connect to the database that stores the LexEVS tables.
- Find the set of tables that stores the ontology using the table prefix.
- The table called codingSchemeSupportedAttrib needs to be updated to disable the hierarchy. To do so, find the row that has the column supportedAttributeTab that has a value called Hierarchy. Update that column to value HierarchyDisabled so that the loaded hierarchy is ignored.
- Restart Tomcat so that the new values are read in and the cache gets cleared.
Systems Administration
How do I increase the Java heap size that Tomcat uses?
- By default we use a 512MB initial heap with a 2GB maximum. You can change these values by editing this file:
/usr/local/tomcat6/conf/tomcat6.conf
- Change the following line to match your requirements. -Xms is initial size, -Xmx is maximum size.
JAVA_OPTS="${DEBUG} -Xms512m -Xmx2G -Djava.awt.headless=true -XX:+CMSClassUnloadingEnabled -XX:MaxPermSize=256m -Djava.net.preferIPv4Stack=true"
- Restart Tomcat by running the following command:
/sbin/service tomcat6 restart
Web User Interface
How can I clear the memcached-based UI cache?
- If you are logged in as the admin user, simply visit http://example/admin and click the "Flush Memcache" button. There should be a response indicating success or failure.
How can I use widgets with my Virtual Appliance?
Working with Javascript-based widgets (Form Autocomplete, Quick Jump/Jump To)
In addition to the existing instructions, you must define an additional Javascript variable in order to have the widgets communicate with your instance of the Virtual Appliance.
var BP_SEARCH_SERVER = "http://{your_appliance_ip_or_domain_name}";
Replace the '{your_appliance_ip_or_domain_name}' text with the IP address or domain name that's assigned to your Virtual Appliance.
Working with Flex/Flash-based widgets (FlexViz, Ontology Tree Widget)
When working with Flash widgets, you'll need to modify the existing instructions to change and add your ip address or domain name. See below and look for '{your_appliance_ip_or_domain_name}' and '{ontology_id}'. The '{ontology_id}' is the numerical id for the ontology you want to use with the widget, sometimes called a virtual id. You can find this via the REST service by looking for the <ontologyId> element, or looking in the Web UI on the Browse page. The links for ontologies contain a number that should be the virtual id.
This example is for the Tree Browser.
<object classid="clsid:D27CDB6E-AE6D-11cf-96B8-444553540000"
id="OntologyTree" width="300" height="100%"
codebase="http://fpdownload.macromedia.com/get/flashplayer/current/swflash.cab">
<param name="movie" value="http://{your_appliance_ip_or_domain_name}/flex/OntologyTree.swf" />
<param name="quality" value="high" />
<param name="bgcolor" value="#ffffff" />
<param name="allowScriptAccess" value="always" />
<param name="flashVars" value="ontology=<%= @ontology.ontologyId%>&alerterrors=false&canchangeontology=false&virtual=true&server=http://{your_appliance_ip_or_domain_name}:8080/bioportal" />
<embed src="http://{your_appliance_ip_or_domain_name}/flex/OntologyTree.swf" quality="high" bgcolor="#ffffff"
width="300" height="100%" name="OntologyTree" align="middle"
play="true"
loop="false"
allowScriptAccess="always"
type="application/x-shockwave-flash"
flashVars="ontology={ontology_id}&alerterrors=false&canchangeontology=false&virtual=true&server=http://{your_appliance_ip_or_domain_name}:8080/bioportal"
pluginspage="http://www.adobe.com/go/getflashplayer">
</embed>
</object>
In addition, there is a bug in existing versions of the Virtual Appliance that is scheduled to be fixed for the .5 version release in March 2012. Until then, you will need to run the following SQL commands to insert records into the 'bioportal' database:
INSERT INTO `ncbo_user` (`id`, `username`, `api_key`, `open_id`, `password`, `email`, `firstname`, `lastname`, `phone`, `date_created`) VALUES (39108, 'annotator_flex', X'32626535656235352D383665622D343164362D386238312D653238653532356163633063', NULL, '8qQ8v8cjUZBCs99voQ2+i4mR1VhyFuj70ZCQUV9caXrPQv4K9FuDIfsfrn+ICoXp\r\n', 'example@domain.org', X'', X'', NULL, '2011-06-28 12:09:32'), (39109, 'search_flex', X'39626437366335382D323230642D346337662D386164302D306135363731313634313939', NULL, '/jj4oWOmI1SXbfczyReBZI9z4DSzRoZP/iX9WJe9lb/7vm1owez/yAh6lIRpNOyj\r\n', 'example@domain.org', X'', X'', NULL, '2011-06-28 12:10:18'), (39110, 'recommender_flex', X'35636564383837322D646636342D343838372D613338652D376338653736333935633830', NULL, 'RBmX8e1X2oUEEcqdp73/X7P7JkrfA7lbK8+svGPEojYmcmxDoHui3gC011Pg1cRF\r\n', 'example@domain.org', X'', X'', NULL, '2011-06-28 12:11:16'), (39111, 'flexviz', X'34363832396130352D386331632D343232622D393530642D313836373633616530663763', NULL, 'uivHblEQJII216fGfG0No4RGELzBtY7Bo14IQulDsQOJY4cLUItYuTtU+TkPs1ln\r\n', 'example@domain.org', X'', X'', NULL, '2011-06-28 12:13:18');