CL:Aligning species-specific anatomy ontologies with CL
The OBO Cell ontology is intended to be a species-neutral reference ontology.
Species-centric anatomy ontologies may want to 'extend' CL with subtypes of cells that are specific to certain types of organism - the discussion on where the dividing line between what goes in a SCA (species-centric anatomy) ontology and CL is still open.
In addition, SCA ontology maintainers may wish to 'replicate' CL terms in their own ontology. Why? The canonical example here is:
Purkinje_cell part_of cerebellum
This assertion is universally true (all Purkinje cells are part_of some cerebellum at all times). The fact that not all organisms have a cerebellum is irrelevant. See RO:part_of for details.
Here is the problem. We have a species-neutral cell ontology with Purkinje_cell (CL:0000121), but we have no species-neutral anatomy ontology with brain parts. We do have for example the zebrafish anatomy ZFA:Main_Page which has ZFA:0000100 cerebellum. So we can say:
CL:0000121 part_of ZFA:0000100
However, this is incorrect as not all Purkinje cells are part of fish cerebellums. Whilst it is not explicitly stated whether the definition of ZFA:0000100 is scoped to any particular organism type, we get into problems if we assume the definitions apply to all organisms in which the type is instantiated, as there will be other links in ZFA that do not hold for all organisms.
If ZFA were to include a term for Purkinje cell, say ZFA:9999999, we could say:
ZFA:999999 part_of ZFA:0000100
SCA curators may have other reasons for replicating CL terms (or terms from other species-neutral reference ontologies like GO) in their ontology.
It's important that SCAs stay in sync with species neutral resources like CL and CARO CARO:Main_Page. This can be done automatically. First the SCA editors must maintain links to the CL; then they can use a reasoner to keep the ontologies in sync.
Maintaining links to CL
The SCA editors have two options here. They can 'explicitly define' SCA specific types in terms of CL types, or they can use a more lightweight xref approach
Explicit definitions
For example, ZFA:Purkinje_cell could be defined as "A CL:Purkinje_cell 'which' is part_of a Zebrafish". This is known as a genus-differentia definition (note: genus is used in its non-linnaean sense here)
This definition must be entered via the oboedit cross-product plugin tool. (The core/generic/genus term would be "CL:Purkinje cell" and the discriminating chacteristics would be "part_of a Zebrafish"). Here is what this would look like in obo 1.2 format:
[Term] id: ZFA:0000134 name: neurons namespace: zebrafish_anatomy xref_analog: ZFIN:ZDB-ANAT-010921-563 intersection_of: CL:0000540 intersection_of: part_of NCBITax:7955 relationship: end ZFS:0000044 ! Adult relationship: part_of ZFA:0000396 ! nervous system relationship: start ZFS:0000026 ! Segmentation:14-19 somites
You will also need a file with the appropriate taxonomic type in it - you can use the whole ncbi_taxonomy.obo file, or you can just keep a separate file with this:
[Term] id: NCBITax:7955 name: Danio rerio
We are now ready to feed this to the reasoner
Using xrefs
The alternate, more lightweight option is to simply maintain xref links to CL. ZFA are doing this anyway (and this part could be automated via simple text-matching). These are "special" xref links in that they have additional implicit meaning. Here is an example of the underlying obo file:
[Term] id: ZFA:0000134 name: neurons namespace: zebrafish_anatomy xref_analog: ZFIN:ZDB-ANAT-010921-563 xref_analog: CL:0000540 relationship: end ZFS:0000044 ! Adult relationship: part_of ZFA:0000396 ! nervous system relationship: start ZFS:0000026 ! Segmentation:14-19 somites
The only difference is the addition of an extra xref line.
This xref isn't enough information for the oboedit reasoner to know that the two ontologies must line up. You must convert the lightweight obo file to an obo file that contains the genus-differentia definitions. This can be done with a simple standalone script:
obo-promote-dbxref-to-intersection.pl --idspace CL -d part_of NCBITax:7955 zfa.obo > zfa_xp.obo
(TODO: add oboedit function to do this)
The script can be downloaded here: http://geneontology.cvs.sourceforge.net/geneontology/go-dev/go-perl/scripts/obo-promote-dbxref-to-intersection.pl?view=log
Here is a section of the output:
[Term] id: ZFA:0000134 name: neurons namespace: zebrafish_anatomy xref_analog: ZFIN:ZDB-ANAT-010921-563 xref_analog: CL:0000540 intersection_of: CL:0000540 intersection_of: part_of NCBITax:7955 relationship: end ZFS:0000044 ! Adult relationship: part_of ZFA:0000396 ! nervous system relationship: start ZFS:0000026 ! Segmentation:14-19 somites
All the script does is add intersection_of lines, which constitute the genus-differentia definition
We can feed the new file (zfa_xp.obo) to the reasoner
Using the reasoner
This section assumes use of the oboedit reasoner. See below for other reasoners
Load up 3 files into oboedit
- zfa_xp.obo (or whatever the SCA is) -- must include intersection_of lines (see above) - cell.obo - zf_taxonomy.obo
See above for the taxonomy file. The only thing that needs to be there is an entry for ZFA:7955. Having the full taxonomy may slow down the reasoner (TODO:test) - however, we may want to try this for projects such as CToL, where we want to reason over multiple related species - but let's keep this simple for now.
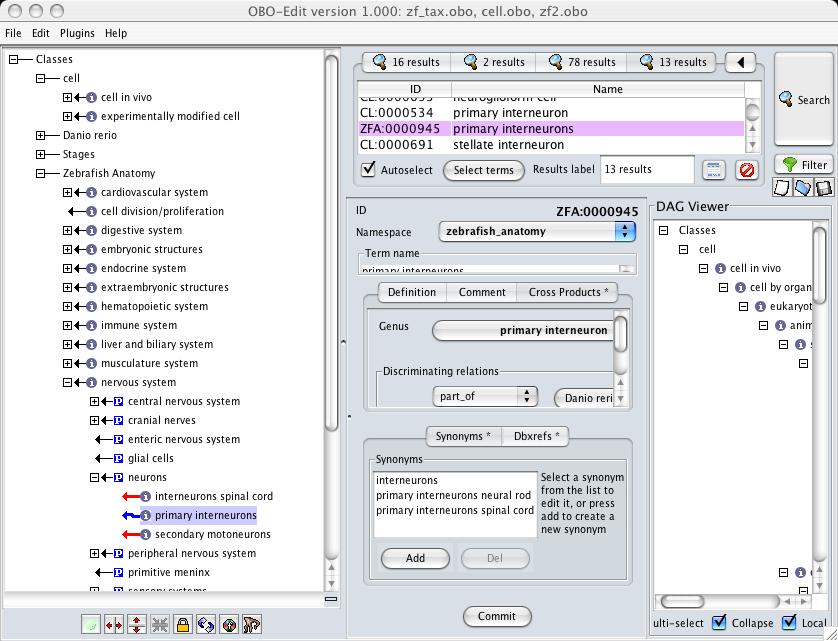
Turn on the reasoner. Exploring the DAG, you will see things like this:
The blue squiggly line between "primary interneurons" and "neurons" indicates the reasoner infers the existence of an (unasserted) is_a link. What should the SCA editor do here? They should make "primary interneurons" an is_a child of "neurons" (remember, although it looks like it is in the display, the blue squiggly line means it is 'inferred'). If the ZFA editor disagrees with this call, they should work with the CL editors to resolve this in CL. The only reason for neither CL or ZFA to amend their ontologies is if the two terms refer to different kinds of entity in reality. If this is the case, the explicit link between the two terms should be removed, and the priniciple of Univocity dictates that either CL or ZFA should change the name of one of their terms.
The straight red line indicates that an 'is_a' link has previously been asserted by the ZFA, and oboedit is showing that had the link not been asserted, it would have been able to figure it out from CL anyway. No action is required on the part of either ZFA or CL editors.
TODO: fill in section on using oboedit queries to find all inconsistencies
TODO: docs on using obo2obo to automoatically fill in is_a links, requiring no interaction on the part of the editor.
Note that the oboedit reasoner is intended for more complicated inferences than simply checking two types that refer to roughly the same entity are in sync (for example, GO processes that refer to CL). However, it works well enough for this simpler use-case.
Pre-reasoned results
Other reasoners
Since most SCAs are edited using oboedit, the oboedit reasoner will be the tool of choice for maintaining consistency between their ontologies and CL/CARO
It is worth noting that other editing tools and reasoners can be used - for example Protege-OWL or SWOOP could be used as the editor, and Pellet or FaCT++ could be used as the reasoner.
To do this with an obo ontology, you'd need to convert to OWL See OboInOWL:Main_Page