Difference between revisions of "NCBO-OOR Server-Side Customization"
Benjamindai (talk | contribs) |
Benjamindai (talk | contribs) |
||
| Line 8: | Line 8: | ||
=BioPortal OntologyLoader Walk-Through= | =BioPortal OntologyLoader Walk-Through= | ||
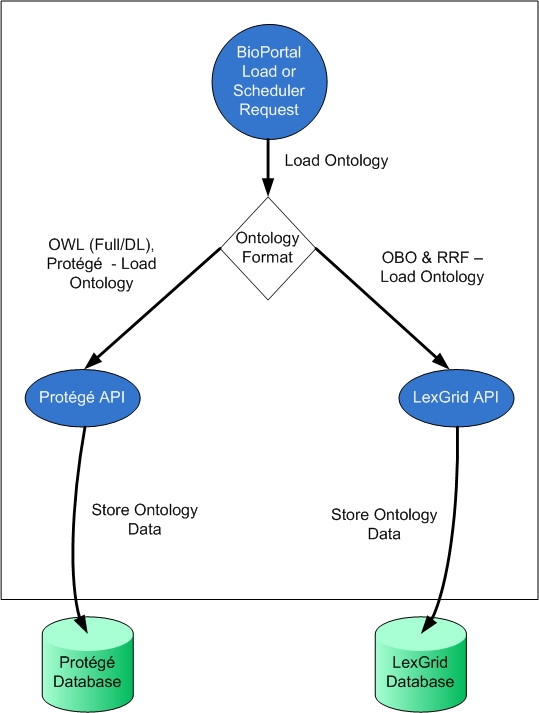
| − | + | Now let's try digging into a more complex example of BioPortal Setter Injection model. When new ontologies are loaded into BioPortal (either through the UI or through back end scheduled jobs), they are loaded either to two possible back-end repositories Protege or LexGrid. All OWL/Protege ontologies are loaded into Protege. All OBO/RRF (which are biomedical specific formats) are loaded into LexGrid. The following diagram presents this general architecture: | |
| + | |||
| + | [[Image:BP Ontology Load Architecture.jpg|center]] | ||
| + | |||
| + | In this scenario, we have the OntologyLoadManager interface which is consumed/used by the OntologyServiceImpl class. Since the OntologyLoadManager is an interface, it has no implementation. | ||
To get my OntologyServiceImpl to accept the injection of the OntologyLoadManager implementations, I define a setting method for that class: | To get my OntologyServiceImpl to accept the injection of the OntologyLoadManager implementations, I define a setting method for that class: | ||
Revision as of 17:41, 10 February 2009
Introduction
NCBO-OOR provides server-side flexibility by leveraging the enterprise pattern of "Dependency Injection." This is accomplished by applying the Spring technology which enables a partner to insert any software implementation that abides to the NCBO-defined interfaces purely by configuration. In other words, a different implementation can be deployed without a recompile or rebuild of the entire NCBO-OOR code-base.
This first NCBO-OOR implemntation is based on the BioPortal architecture and code-base. Thus, the majority of the developer considerations for NCBO-OOR is generally applicable to the BioPortal (and vice-versa). The author will note differences where appropriate.
BioPortal Server-Side Pattern: Dependency Injection
The gist of dependency injection is that a seperate object, an assembler, populates the implementation of a defined interface that can be used by any consuming object that understands the interface. Spring implements a specific variation of this patterns called "Setter Injection." A significant number of BioPortal server-side capabilities heavily leverage this pattern. This approach enables these BioPortal capabilities to easily swap out different implementations. To learn more about this pattern in general, please see [Martin Fowler's excellent exposition on this topic | http://martinfowler.com/articles/injection.html]. The following sections elucidate a couple BioPortal-specific examples.
BioPortal OntologyLoader Walk-Through
Now let's try digging into a more complex example of BioPortal Setter Injection model. When new ontologies are loaded into BioPortal (either through the UI or through back end scheduled jobs), they are loaded either to two possible back-end repositories Protege or LexGrid. All OWL/Protege ontologies are loaded into Protege. All OBO/RRF (which are biomedical specific formats) are loaded into LexGrid. The following diagram presents this general architecture:
In this scenario, we have the OntologyLoadManager interface which is consumed/used by the OntologyServiceImpl class. Since the OntologyLoadManager is an interface, it has no implementation.
To get my OntologyServiceImpl to accept the injection of the OntologyLoadManager implementations, I define a setting method for that class:
class OntologyServiceImpl...
Map<String, OntologyRetrievalManager> ontologyRetrievalHandlerMap
public void setOntologyLoadHandlerMap(Map<String, OntologyLoadManager> ontologyLoadHandlerMap)
this.finder = finder;
}
One then specifies in the BioPortal Spring configuration file ("applicationContext-services.xml") the implementations to inject into OntologyServiceImpl. I've appended an excerpt from the configuration file:
<bean id="conceptService"
class="org.ncbo.stanford.service.concept.impl.ConceptServiceImpl">
<property name="ncboOntologyVersionDAO">
Cite error: Invalid <ref> tag; invalid names, e.g. too many
</property>
<property name="ontologyFormatHandlerMap">
Cite error: Invalid <ref> tag; invalid names, e.g. too many
</property>
<property name="ontologyRetrievalHandlerMap">
Cite error: Invalid <ref> tag; invalid names, e.g. too many
</property>
</bean>
The above configuration specifies that "ConceptServiceImpl" has a setter method named "setOntologyFormatHandlerMap" which accepts an injection of the map named "ontologyFormatHandlerMap" which includes both "OntologyLoadManagerProtegeImpl" and "OntologyLoadManagerLexGridImpl" as implementations of "OntologyLoadManager."