Search results
Jump to navigation
Jump to search
Page title matches
- #REDIRECT [[PATO:Annotations]]30 bytes (3 words) - 15:11, 15 December 2008

File:Human annotations.zip GO Annotations for human (Homo sapiens).(1.56 MB) - 10:01, 27 March 2007
File:Yeast annotations.zip GO annotations for yeast (Saccharomyces cerevisiae).(1.02 MB) - 10:00, 27 March 2007
Page text matches
- * All annotations in OBD-Phenotype subset * All annotations to entities in OMIM IDspace850 bytes (126 words) - 14:34, 5 March 2008
- ...pabilities for computing the similarity between entities based on ontology annotations. OBD is flexible with respect to what kind of annotations and ontologies are used. For example:1 KB (196 words) - 13:00, 9 October 2008
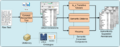
File:OBA service workflow.png ...and NCBO ontologies. Second, different components expand the first set of annotations using ontologies semantics.(1,057 × 414 (133 KB)) - 09:22, 20 October 2009- #REDIRECT [[PATO:Annotations]]30 bytes (3 words) - 15:11, 15 December 2008

File:Yeast annotations.zip GO annotations for yeast (Saccharomyces cerevisiae).(1.02 MB) - 10:00, 27 March 2007
File:Human annotations.zip GO Annotations for human (Homo sapiens).(1.56 MB) - 10:01, 27 March 2007- ...OWL, download the ''Saccharomyces cerevisiae'' or the ''Homo sapiens'' GO Annotations in OWL. Both files import the GO ontology in OWL, so you have to have it in [[Media:Yeast_annotations.zip|Download Yeast GO Annotations in OWL]]3 KB (496 words) - 17:20, 3 April 2007
- ...ssemblies and compute pipelines (the results of which are sometimes called annotations, although the word is typically only used for human-curated data). As well2 KB (318 words) - 16:15, 15 May 2006
- ...y the OBA/OBR workflow. It contains the concepts and terms used for direct annotations as well as the relations between concepts used during the semantic expansio1 KB (162 words) - 10:27, 18 September 2009
- conceptNodes <- getNodeSet(x,"/success/data/annotatorResultBean/annotations/annotationBean/concept") resultNodes <- getNodeSet(x, "/success/data/annotatorResultBean/annotations/annotationBean/context")3 KB (279 words) - 10:41, 19 December 2013
- * add new types of annotations1 KB (172 words) - 10:53, 17 October 2008
- The Sequence Ontology: A tool for the unification of genome annotations. <BR>Eilbeck K., Lewis S.E., Mungall C.J., Yandell M., Stein L., Durbin R.,1 KB (219 words) - 18:55, 25 January 2007
- '/success/data/annotatorResultBean/annotations/annotationBean/concept'); '/success/data/annotatorResultBean/annotations/annotationBean/context');3 KB (415 words) - 09:00, 15 September 2011
- '''Minimal requirement for term annotations in the CTO (metadata)'''2 KB (279 words) - 08:35, 15 March 2007
- '''Minimal requirement for term annotations in the CTO (metadata)'''2 KB (285 words) - 11:58, 6 July 2007
- ''To automatically recommend an ontology to use for semantic annotations'' ...tic annotation based approach and scores the ontologies according to those annotations. The recommender service uses the [[Annotator Web service]] . The prototype4 KB (489 words) - 13:28, 12 September 2011
- ...) of full page text articles, for which you wish to retrieve and parse the annotations. Assuming it takes about 20 second to parse and organize the response for e ...r BioPortal users. If the constraints suggested by this article mean your annotations will take too long, then you will need to set up a BioPortal Virtual Applia8 KB (1,421 words) - 21:15, 7 February 2017
- ...rge repository of biomedical ontologies, terminologies, and ontology-based annotations. BioPortal is available at: http://bioportal.bioontology.org/. All content ...ab to search for term annotations or from the Resource Index tab to browse annotations for an individual term. This tool enables researchers to search for biomedi11 KB (1,718 words) - 18:15, 7 June 2022
- ...iption) an annotation has been produced. This information is used to score annotations.5 KB (669 words) - 19:16, 8 April 2013
- ...wn to a location on sequence, and be able to reason over the parts of gene annotations. To define the term gene in SO, we asked several questions. What is the ext3 KB (512 words) - 09:56, 10 August 2006